Neuronal Maturation in the Developing Human Brain
Post by Shireen Parimoo
What's the science?
Ribonucleic acid (RNA) sequencing is a technique that is often used to create a genetic profile of cells in the brain. When applied at the level of single cells, RNA sequencing can be used to discern their identity. Studies using RNA sequencing have recently shown that many different types of neurons in the developing human brain arise from radial glia and other progenitor cells (cells that can differentiate into other cells). Although genetic profiles are useful for identifying cell types, other factors like cell physiology and morphology can also provide insight into cell identity. Currently, the physiological properties of various cells in the developing human brain – like how they respond to neurotransmitters – are not known. This week in Neuron, Mayer and colleagues developed a novel technique to combine RNA sequencing with cell imaging measures to identify the genetic and physiological profiles of developing human neocortical cells.
How did they do it?
The authors identified different cell types in tissue samples from the first and second trimester of the developing human neocortex including ventricular radial glia, outer radial glia, intermediate progenitor cells, and newborn neurons. They analyzed an RNA sequencing dataset to identify the gene expression levels of various neurotransmitter receptors and receptor subunits. They used single-molecule fluorescent in-situ hybridization and immunohistochemistry to examine the expression of a serotonergic receptor (HTR2A) and a purinergic receptor (P2RY1) in the radial glia specifically. To investigate the electrophysiological properties of these cells, they applied receptor agonists (molecules that bind to and activate receptors) to tissue slices and measured the change in the resulting electrical currents. Using calcium imaging as a measure of activity, they examined the effect of agonists on single cells from various regions of the neocortical tissue. To map the genetic profiles of neocortical cells with their physiological response profiles, the authors dissociated cells and dosed them with various receptor agonists to record their response, followed by RNA sequencing. They then investigated whether the maturational stage of newborn neurons affects their physiological responses. Finally, they trained two types of machine learning models (a supervised Bayes classifier and an unsupervised classifier) to predict cell identity from their genetic and physiological response profiles.
What did they find?
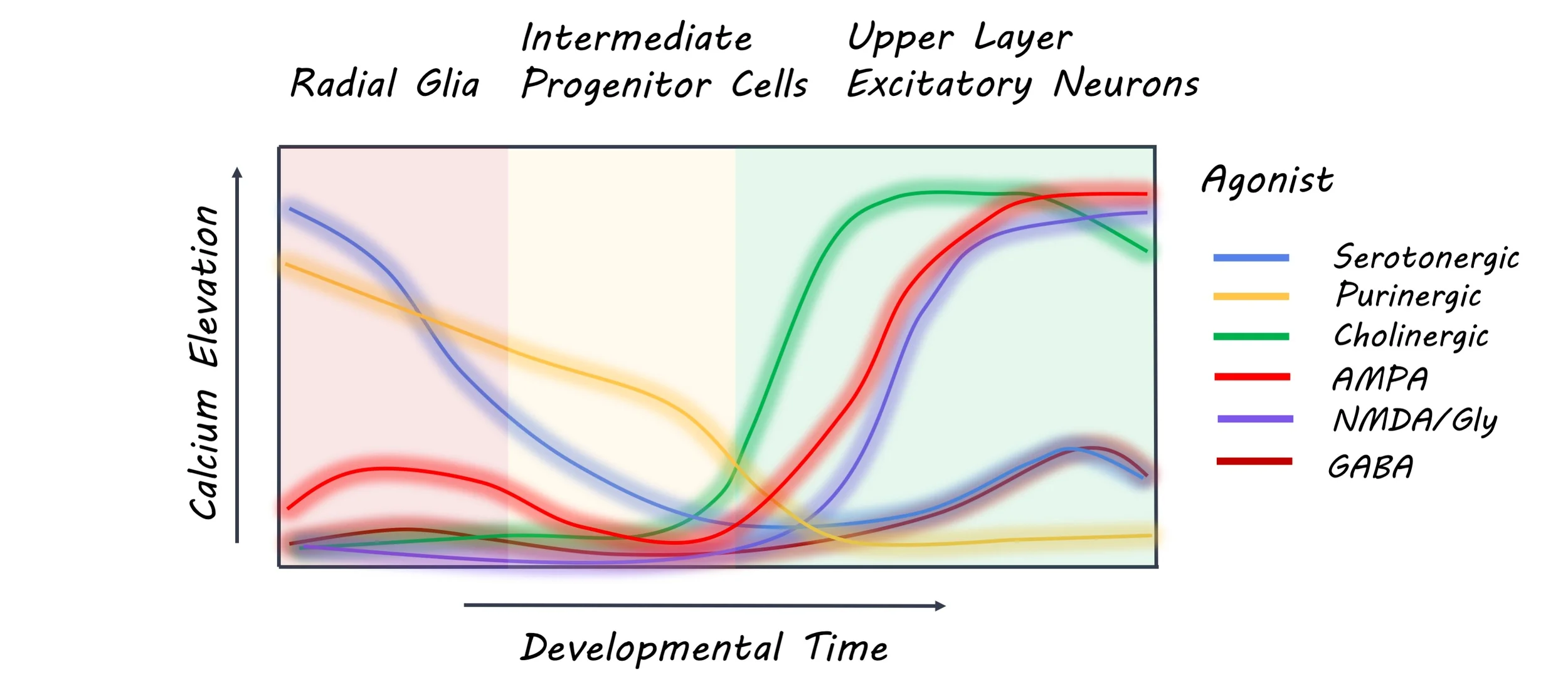
Cells in the developing brain differentially expressed genes for neurotransmitter receptors and their subunits. For example, the GRIN2A subunit of the NMDA receptor was highly expressed in progenitor cells, whereas the GRIN2B subunit was only expressed in neurons. Similarly, there was a high expression of the HTR2A receptor in radial glial cells. Application of receptor agonists also revealed a distinct response profile for each cell type. Specifically, NMDA receptors induced currents with different properties in neurons and radial glia. As the HTR2A receptor expression was only observed in the radial glial cells, applying agonists only induced currents in the ventricular and outer radial glia. In fact, applying an HTR2A antagonist (inhibitor) disrupted the morphology of the outer radial glia after 72 hours. This means that the various cells in the developing brain not only have distinct genetic and physiological response profiles, but these properties may also affect their morphology.
Neurons had more heterogeneous response profiles than progenitor cells, and this varied by their stage of maturation. The Bayes classifier was able to predict cell identities (in terms of physiological properties) based on genetic data. However, it identified some cells as immature neurons, even though their physiological responses were more similar to mature neurons. Therefore, different readouts may help to better define neuronal maturation. On the other hand, the unsupervised classifier clustered cell types based on their physiological response profiles. Although genetic and physiological identification generally matched, some genetically identified cell types had several possible physiological responses, which was related to their maturational stage, for instance. This means that the same neuron can respond differentially to neurotransmitter signaling depending on its stage of maturation.
What's the impact?
This is the first study to use multimodal analyses to map the genetic profiles of developing human neocortical cells to their physiological profiles. In particular, the finding of distinct physiological response profiles across cell types is important because it highlights how the function of various cells and neurons in the brain can change as they mature. This has important implications for understanding the functional role of various cells and neurons in the brain.
Mayer et al. Multimodal single-cell analysis reveals physiological maturation in the developing human neocortex. Neuron. (2019). Access the original scientific publication here.